Impact of Downsampling Size and Interpretation Methods on Diagnostic Accuracy in Deep Learning Model for Breast Cancer Using Digital Breast Tomosynthesis Images
Ryusei Inamori
Tohoku University
Tomofumi Kaneno
Tohoku University
Ken Oba
St. Luke’s International Hospital
Eichi Takaya
Tohoku University
Daisuke Hirahara
Harada Academy
Tomoya Kobayashi
Tohoku University
Kurara Kawaguchi
Tohoku University
Maki Adachi
Tohoku University
Daiki Shimokawa
Tohoku University
Kengo Takahashi
Tohoku University
Hiroko Tsunoda
St. Luke’s International Hospital
Takuya Ueda
Tohoku University
The Tohoku Journal of Experimental Medicine
Abstract
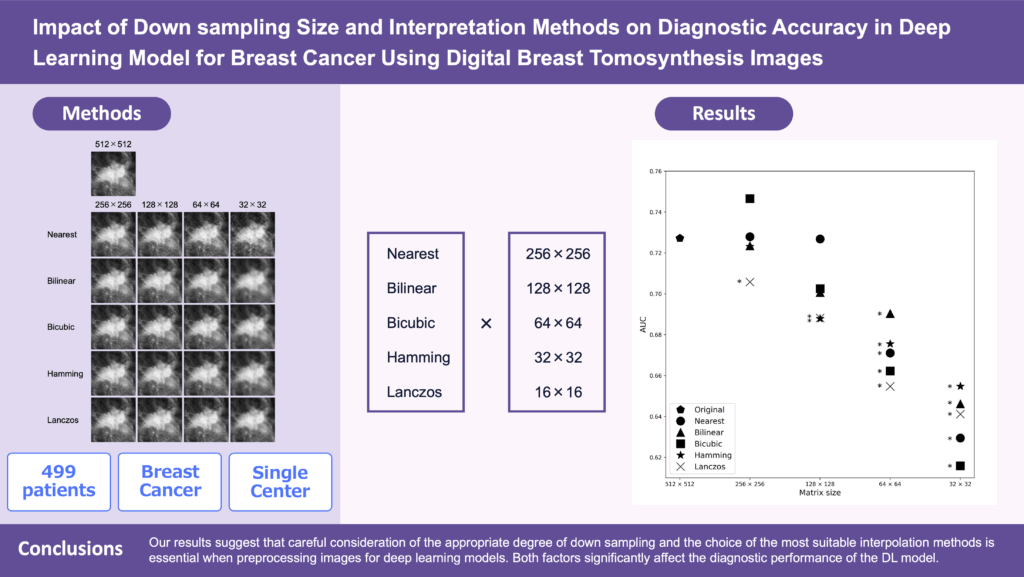
While deep learning (DL) models have shown promise in breast cancer diagnosis using digital breast tomosynthesis (DBT) images, the impact of varying matrix sizes and image interpolation methods on diagnostic accuracy remains unclear. Understanding these effects is essential to optimize preprocessing steps for DL models, which can lead to more efficient training processes, improved diagnostic accuracy, and better utilization of computational resources. Our institutional review board approved this retrospective study and waived the requirement for informed consent from the patients. In this study, 499 patients (29–90 years old, mean age 50.5 years) who underwent breast tomosynthesis were included. We performed downsampling to 256×256, 128×128, 64×64, and 32×32 using five image interpolation methods: Nearest (NN), Bilinear (BL), Bicubic (BC), Hamming (HM), and Lanczos (LC). The diagnostic accuracy of the DL model was assessed by mean AUC with its 95% confidence interval (CI). DL models with downsampled images to 256×256 pixels using the LC interpolation method showed a significantly lower AUC than the original 512×512 pixels model. This decrease was also observed with the 128×128 pixels DL models using HM and LC methods. All interpolation methods showed a significant decrease in AUC for the 64×64 and 32×32 pixels DL models. Our results highlight the significant impact of downsampling size and interpolation methods on the diagnostic performance of DL models. Understanding these effects is essential for optimizing preprocessing steps, which can enhance the accuracy and reliability of breast cancer diagnosis using DBT images.
Method
Images were cropped to 512×512 pixels centered on radiologist annotations (or randomly for normal tissue) and shifted if overlapping the DBT boundary. Each image was downsampled to 256, 128, 64, and 32 pixels using five interpolation methods (Nearest, Bilinear, Bicubic, Hamming, Lanczos) from the Python Pillow library. Figure 1 shows examples of these downsampled images (using CMMD dataset: mammography).
Results
The original 512×512 DL model achieved an AUC of 0.727 (95% CI: 0.712–0.742). At 256×256, Lanczos interpolation reduced AUC significantly; at 128×128, Hamming and Lanczos also decreased performance. All methods at 64×64 and 32×32 showed significantly lower AUCs than the original (Figure 2, Table 1).
Citation
If you find our work or any of our materials useful, please cite our paper:
@article{Ryusei Inamori20252024.J071,
title={Impact of Downsampling Size and Interpretation Methods on Diagnostic Accuracy in Deep Learning Model for Breast Cancer Using Digital Breast Tomosynthesis Images},
author={Ryusei Inamori and Tomofumi Kaneno and Ken Oba and Eichi Takaya and Daisuke Hirahara and Tomoya Kobayashi and Kurara Kawaguchi and Maki Adachi and Daiki Shimokawa and Kengo Takahashi and Hiroko Tsunoda and Takuya Ueda},
journal={The Tohoku Journal of Experimental Medicine},
volume={265},
number={1},
pages={1-6},
year={2025},
doi={10.1620/tjem.2024.J071}
}